Scientists Invented AI Made From DNA
Researchers made a neural network out of DNA that can recognize handwritten numbers
In July (2018), researchers at Caltech announced that they created an artificial neural network from synthetic DNA that is able to recognize numbers coded in molecules. It’s a novel implementation of a classic machine learning test that demonstrates how the very building blocks of life can be harnessed as a computer.
This is pretty mind blowing, but what does it all mean? For starters, “artificial intelligence” here doesn’t refer to the superhuman AI that is so beloved by Hollywood. Instead, it refers to machine learning, a narrow form of artificial intelligence that is best summarized as the art and science of pattern recognition. Most of the cutting edge advances in machine learning involve artificial neural networks, which are a type of computing architecture loosely based on the human brain. These neural networks are fed a lot of data as input and then taught how to perform some task with that data; sometimes humans help to guide the algorithm’s learning, and sometimes not.
This is effectively what the Caltech researchers designed, but instead of using silicon and transistors, they used DNA and test tubes as their neural network’s hardware.
All DNA is composed of four basic nucleotides: adenine (A), cytosine (C), guanine (G), and thymine (T). Strands of these nucleotides can bond with other strands of these nucleotides to form the double helix of DNA, but can only bind in specific combinations (i.e., A-T or C-G). This predictable pattern of combination makes these nucleotide strands ideal computing devices, which can be designed so that they produce specific chemical reactions in the presence of various molecules.
The Caltech researchers applied this sort of DNA-based computer to one of the classic tests in computer vision research: teaching an algorithm how to recognize handwritten numbers. This is tough for a computer to do because humans all write the number four slightly differently. Humans are hardwired to easily see the similarities between the ways different people write four, but machines don’t have such biological luxuries. By feeding an artificial neural network a ton of handwritten examples of the number four, however, an algorithm can “learn” to generalize qualities from individual examples and form an abstract idea of what a written four looks like. The next time the algorithm encounters something that looks like a four, it will compare this to its abstract representation of four and if it’s a close enough match, it will conclude that it is looking at a four.
In 2011, Caltech bioengineer Lulu Qian created the first artificial neural network out of DNA, but it could recognize only a handful of patterns. In the work unveiled recently, one of Qian’s graduate students, Kevin Cherry, has considerably advanced this technique by applying it to the recognition of handwritten “molecular numbers”. Each molecular number was based on a handwritten number translated into a 20-bit pattern in a 100-bit (10×10) grid. Each of the bits on the grid was represented by a molecule of DNA, and these molecules of DNA were assigned a place on a conceptual 10×10 grid before being mixed together in a test tube.
The DNA in the test tube doesn’t resemble a grid – it’s all mixed up – and so a molecule’s place on the grid was determined by the concentration of each molecule in the test tube. The DNA neural net was a strand of DNA that produced a specified reaction when added to the test tube only if the 20 DNA molecules assigned to represent a given number are arranged (i.e., in the appropriate concentrations) so that they form that number when translated onto the 10×10 grid.
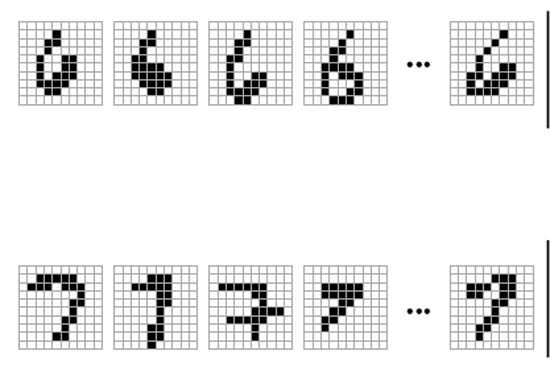
Examples of handwritten sixes and sevens arranged on a 10×10 grid. Each square on the grid is represented by a molecule in a specified concentration.
Cherry began his experiment by building a neural net that could distinguish between handwritten sixes and sevens that had been translated into molecular structures. He tested this approach on 36 different handwritten versions of the same numbers and in each instance the DNA neural network was able to recognize them. Cherry used a “winner take all” approach to allow DNA neural nets to distinguish between numbers by synthesizing a so-called “annihilator” molecule.
“The annihilator forms a complex with one molecule from one competitor and one molecule from a different competitor and reacts to form inert, unreactive species” Cherry said. “The annihilator quickly eats up all of the competitor molecules until only a single competitor species remains. The winning competitor is then restored to a high concentration and produces a fluorescent signal indicating the networks’ decision.”
Importantly, this winner take all approach also allowed the DNA neural net to differentiate between the numbers 1-9 in a DNA soup. After undergoing its reactions, the test tube would show two fluorescent signals, which would indicate which number was represented in the test tube. For example, green and yellow fluorescence represented a five, and green or red represented a nine.
Looking to the future, Cherry and Qian hope that this technique can be augmented by adding memory functions to their DNA neural nets and allow for improved medical testing.
“Common medical diagnostics detect the presence of a few biomolecules, for example cholesterol or blood glucose.” Cherry said. “Using more sophisticated biomolecular circuits like ours, diagnostic testing could one day include hundreds of biomolecules, with the analysis and response conducted directly in the molecular environment.”
yogaesoteric
October 11, 2018
Also available in:
 Français
Français
